DiffModeler Set to Improve Protein Structure Modeling
11-01-2024

A new tool developed at Purdue University, called DiffModeler, is poised to transform how scientists model complex protein structures.
This innovation holds great promise for advancing our understanding of molecular biology and accelerating breakthroughs in fields like drug discovery.
Spearheaded by Professor Daisuke Kihara, an interdisciplinary team from Purdue’s Biological Sciences and Computer Science departments has created an automated method that significantly improves the accuracy of protein modeling, particularly when using cryogenic electron microscopy (cryo-EM) at challenging resolution levels. They published their findings in Nature Methods.
Challenges in Protein Structure Modeling
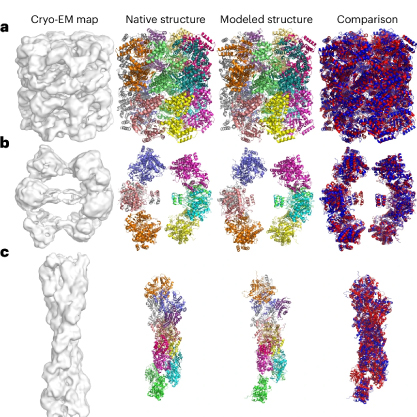
Proteins are essential molecules responsible for various functions in living organisms, including catalyzing chemical reactions, signaling within and between cells, and transporting molecules. To understand how these processes work, researchers need to determine the three-dimensional (3D) structure of proteins, particularly in large, multi-chain complexes. Cryo-EM, a technique that allows researchers to capture images of these protein complexes in their native state, has grown in popularity. However, when the resolution of cryo-EM images falls in the intermediate range (around5 to 10 Ångströms), determining protein structures becomes difficult due to the limited detail in the images.
At these intermediate resolutions, conventional tools often fail to trace protein backbones accurately or identify the positions of amino acids, which are the building blocks of proteins. This is where DiffModeler steps in.
Introducing DiffModeler
DiffModeler is a fully automated system designed to model large protein complexes from cryo-EM maps in these challenging resolution ranges. It uses an innovative diffusion model to enhance the clarity of the protein backbones in the maps, making it easier to fit predicted protein structures into the cryo-EM data.
In addition to the diffusion model, DiffModeler integrates AlphaFold2, a protein prediction tool that predicts the structures of individual protein chains, which won the Nobel Prize in Chemistry this year. By combining these predicted single-chain structures with enhanced cryo-EM data, DiffModeler assembles highly accurate models of entire protein complexes, even in cases where previous methods had fallen short.
Testing of DiffModeler has shown remarkable results. The tool achieved an average TM-score of 0.92 in its modeling efforts, a significant improvement over other existing methods. The TM-score, a widely used metric for comparing the accuracy of protein structure models, ranges from 0 to 1, with values closer to 1 indicating higher accuracy.
"Diffmodeler combines the strengths of AlphaFold and a recent advanced AI approach, the diffusion model,” explains Kihara. “Enabling researchers to obtain protein structure models even at low map resolutions, which have traditionally been challenging to model.”
Interdisciplinary Collaboration
The development of DiffModeler is the result of collaboration across multiple disciplines, showcasing Purdue’s strength in interdisciplinary research. Professor Kihara's expertise spans both biological sciences and computer science, and the project also involved contributions from researchers in computer science and engineering at Purdue, as well as collaborators from the Vellore Institute of Technology in India. Together, the team combined computational modeling, machine learning, and biological science to create a tool that is set to advance our understanding of complex biological systems.
Future Applications and Impact
DiffModeler’s potential impact is vast. It opens new possibilities for studying protein structures that were previously too difficult to model, which is especially important for the fields of drug discovery and molecular biology. Accurately modeled protein structures can reveal binding sites for potential drug compounds, aiding in the design of new therapies for diseases.
Moreover, DiffModeler can be used in studying how proteins interact with DNA and RNA, offering insights into the molecular mechanisms behind genetic expression and regulation. Its ability to model not just proteins, but also protein-DNA/RNA complexes, is a testament to its versatility.
The successful application of DiffModeler in challenging cases, such as modeling protein complexes with up to 47 chains and more than 13,000 residues, highlights its capability to handle large and intricate structures, which are often seen in real biological systems. These successes make it a valuable tool for structural biologists.
About the Department of Biological Sciences at Purdue University
Purdue Biological Sciences is the largest department in the Life Sciences at Purdue University. As part of Purdue One Health, we are dedicated to pioneering scientific discoveries and transformative education at the cutting edge of innovation. From molecules to cells, from tissues to organisms, from populations to ecosystems - we bring together multiple perspectives, integrating across biological scales to advance our understanding of life and tackle the world’s most pressing challenges. Learn more at bio.purdue.edu/.
Writer: Alisha Willett, areferda@purdue.edu
Source: Daisuke Kihara, dkihara@Purdue.edu